Project 1
Apoptotic cell clearance during development
During normal development of multicellular organisms, a large number of cells is eliminated through apoptosis and subsequent phagocytosis by “professional” macrophages and immature dendritic cells and “non-professional” tissue neighboring cells. Drosophila phagocytic cells, professional macrophages and non-professional glia, are highly homologous, both functionally and molecularly, to their mammalian counterparts. Yet, phagocytosis in flies is less redundant than in vertebrates, which allows studying its molecular mechanisms in great detail. We use Drosophila embryogenesis and metamorphosis to study the molecular mechanisms of apoptotic cell clearance during development. Apoptotic cell clearance is a dynamic process which proceeds in four main steps: (1) recruitment of phagocytes by the apoptotic cell, (2) recognition of the cell as a target for phagocytosis, (3) engulfment of the targeted cell, and (4) phagosome maturation and degradation of the apoptotic cell. We focus on the role of transmembrane receptors, intracellular signaling and cytoskeletal rearrangements during distinct steps of the process.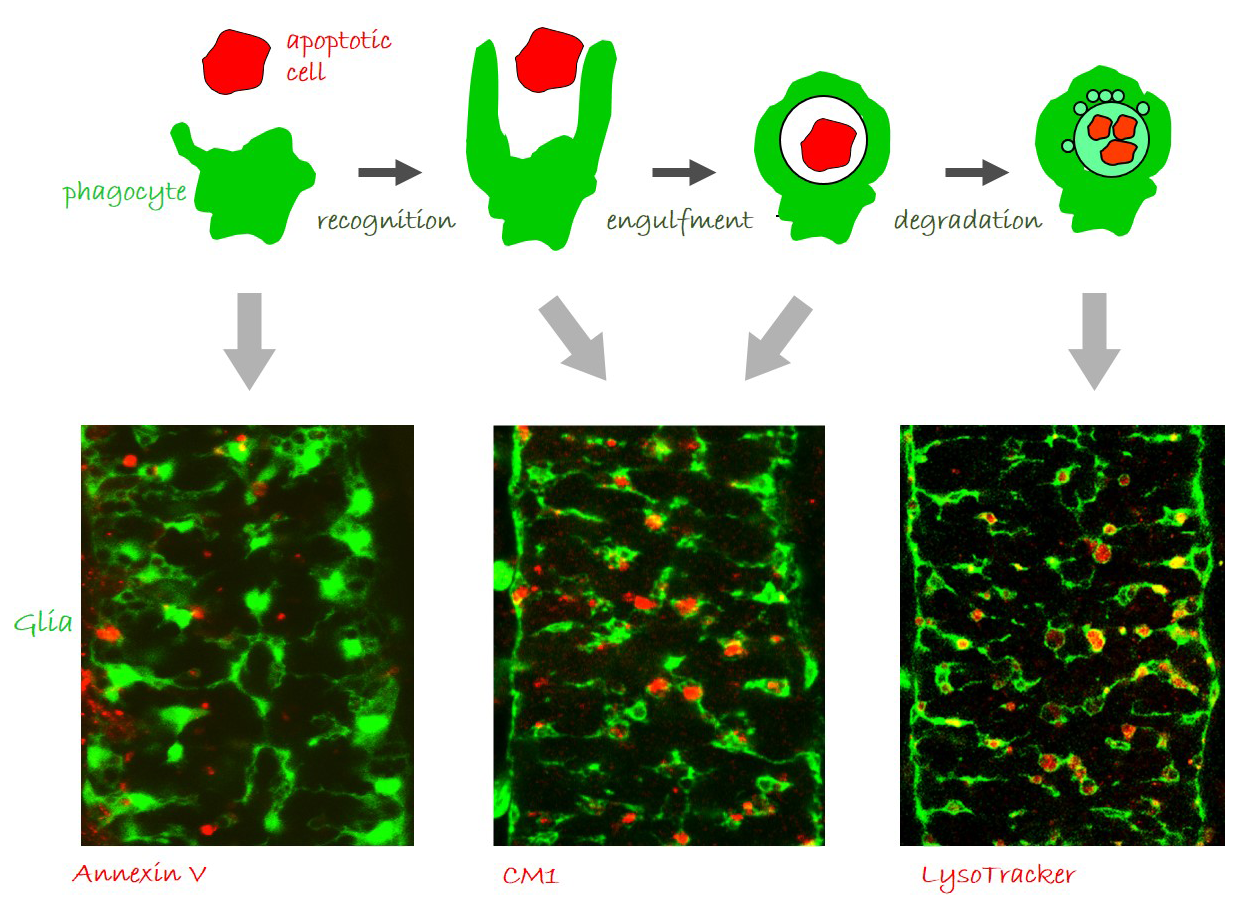
Glia perform phagocytosis in the embryonic CNS.
(A) Schematic depicting the different stages of phagocytosis – apoptotic cell recognition, engulfment, phagosome formation and maturation. (B-D) Projections from confocal stacks of the embryonic CNS at stage 16, ventral view, showing GFP-labeled glia and different apoptosis/ phagocytosis markers in red: (B) Annexin V, (C) anti-activated Caspase-3 antibody, and (D) LysoTracker. Bar, 20 µm.
Project 2
Establishment of phagocytic ability
Phagocytes must recognize apoptotic cells with high level of specificity in order to remove apoptotic but not living cells. This very precise discrimination is achieved through transmembrane phagocytic receptors or secreted bridging molecules, which recognize specific targets on apoptotic surfaces. Importantly, most of the receptors are exclusively expressed in phagocytic cell populations. We are interested in molecular mechanisms that control the establishment of phagocytic ability in developing glia and macrophages. Our studies demonstrate that glial phagocytic ability is determined by a developmental program responsible for the specific expression and function of two glial phagocytic receptors, Six-Microns-Under (SIMU) and Draper. Undifferentiated glial cells in repo mutant embryos exhibit abnormal expression of SIMU and Draper resulting in abnormal phagocytosis of apoptotic cells. Current research focuses on molecular mechanisms governing establishment of phagocytic ability in embryonic macrophages.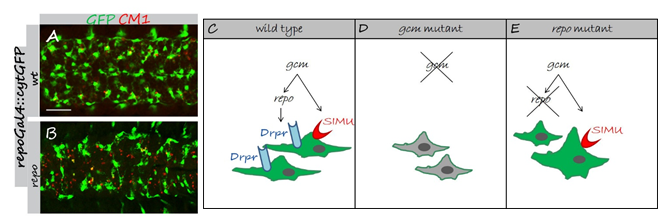
Developmental regulation of glial phagocytic ability during embryogenesis.
(A,B) Projections from confocal stacks of the CNS at embryonic stage 16, ventral view; apoptotic cells are in red (CM1) and glia in green (repoGal4::cytGFP). Bar, 20 µm. In wild type embryo (A) apoptotic particles are mostly inside GFP-positive glia. In repo mutant embryo (B) many apoptotic particles are outside GFP-positive glia. (C-E) Schematic summary of developmental regulation on SIMU and Draper expression in embryonic glia. (C) Wild type. (D) In gcm mutant no glia are formed and no SIMU and Draper expression is found in cells, which turn from glia to neurons. (E) repo mutant glia appear in abnormal shapes. SIMU is expressed on some glial cells, which are often bigger than wild type glia and show macrophage-like behavior.
Project 3
The role of phagocytic glia in aging and neurodegeneration
Recent studies in both vertebrates and invertebrates showed that neurons interact with glia from very early stages of development and that these interactions are critical for the establishment, function and maintenance of the nervous system.We take advantage of the fly model in order to elucidate molecular mechanisms underlying interactions between neurons and phagocytic glia during aging and neurodegeneration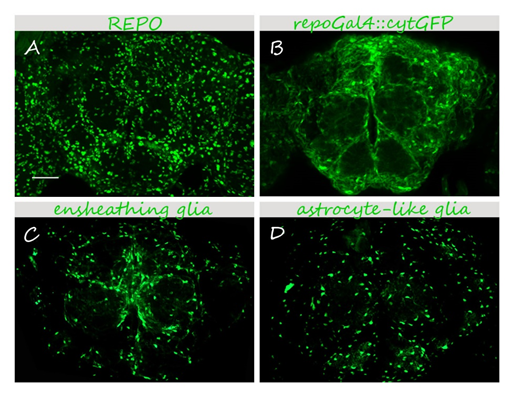
Glia in the adult Drosophila brain.
25-40 μm 3D projections are shown. (A) anti-REPO immunostaining labels all glial nuclei. (B-D) Glial cells are visualized using specific drivers and a cytoplasmic GFP reporter (UAScytGFP). (B) pan-glial <em>repoGal4 </em>labels all glial cells. (C,D) Specific drivers for ensheathing (C) and astrocyte-like (D) glia depict specific glial cell types. Bar, 50 μm
